Fluorescence in situ hybridization (FISH) is a powerful molecular biology technique that allows us to visualize and map the location of specific DNA or RNA sequences in cells and tissues. It has contributed significantly to our understanding of genetics, gene expression, chromosomal abnormalities, and various biological processes.
The history of FISH is marked by important developments and milestones:
1. Early concepts and developments (1960s and 1970s)
- The foundation for FISH was laid in the 1960s when researchers began experimenting with nucleic acid hybridization techniques. Hybridization involves linking complementary strands of DNA or RNA.
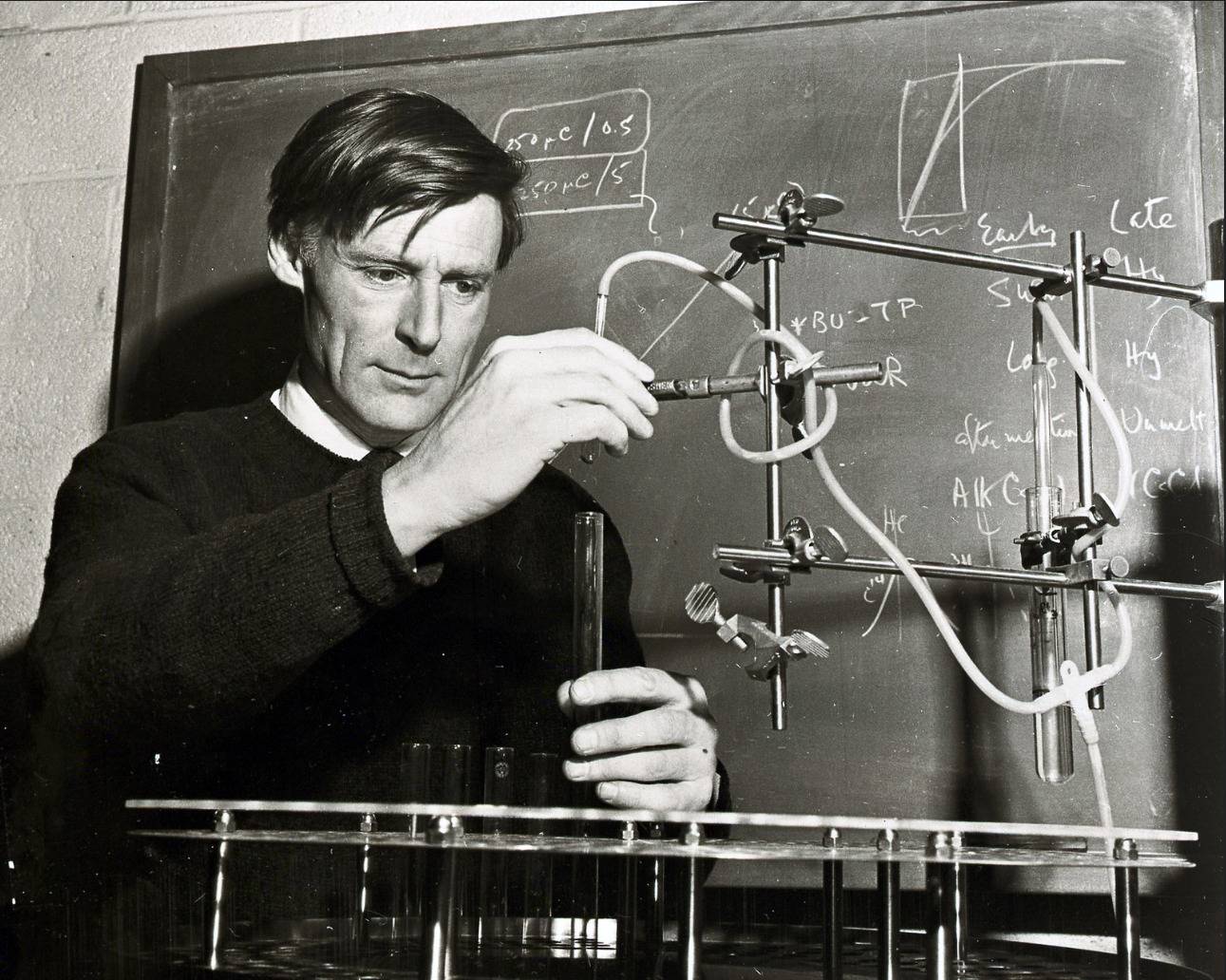
- In 1969, John Cairns (see photo) used radioactive probes to detect DNA sequences in the bacterial genome, which was one of the first cases of in situ hybridization.
- In the early 1970s, researchers such as Mary-Lou Pardue and Joseph Gall used radioactive RNA probes to visualize specific gene sequences on Drosophila (fruit fly) chromosomes.
2. Introduction of non-radioactive probes (1980s)
- The use of radioactive probes raised concerns about user safety and disposal of the material, leading to the development of non-radioactive probes for FISH.
- In 1980, Louise Brown and John Smith published a method using biotin-labeled probes for in situ hybridization.
- During the 1980s, researchers investigated alternative labeling techniques, such as digoxigenin-labeled probes, which produced detectable signals without using radioactivity.
3. Fluorescent labeling and confocal microscopy (1980s and 1990s)
- The breakthrough for FISH came with the integration of fluorescent dyes as labels for FISH probes. This enabled easier detection and visualization of hybridization events.
- In 1986, John W. Sedat and David A. Agard introduced wide-field epifluorescence microscopy to visualize FISH probes.
- In 1989, Robert Singer and colleagues developed the method of fluorescence in situ hybridization using directly labeled DNA probes.
4. Chromosome painting and spectral karyotyping (SKY) (1990s–2000s)
- Researchers began using multiple fluorescent probes to label different chromosomes simultaneously, a technique known as chromosome painting.
- In 1995, the Spectral Karyotyping (SKY) technique was introduced, which allowed the simultaneous visualization of all chromosomes using different fluorescent colors.
- These techniques revolutionized the study of chromosomal abnormalities and genetic variations.
5. Advances in probe design and automation (2000-present)
- FISH has found applications in cancer diagnostics, prenatal testing, and several other areas of research.
- Biotrack ( the parent company of NL-Lab ) develops C-FISH, a patented automated version of FISH that uses powerful computers, high-resolution cameras and AI-driven algorithms.
- The Biotrack platform, based on C-FISH technology, enables efficient analysis of large numbers of samples, even for complex applications (such as gut microbiome research).
- Biotrack is continuously innovating in probe design, fluorescent labeling technologies, digital image processing, and AI-driven algorithms to further improve the unprecedented sensitivity, specificity, and speed of FISH.
The history of FISH illustrates the evolution of molecular techniques and their profound impact on our understanding of genetics and biology. FISH remains a fundamental tool for studying genome organization, gene expression, and chromosomal abnormalities in various organisms and cell types.
Interested or have questions?
If you have any questions following this article, we strongly recommend that you contact us .